Saccharomycotina
Saccharomycetales
Meredith Blackwell, Cletus P. Kurtzman, Marc-André Lachance, and Sung-Oui Suh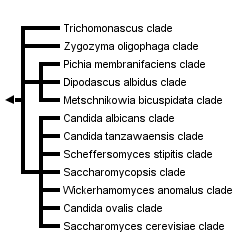

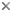
This tree diagram shows the relationships between several groups of organisms.
The root of the current tree connects the organisms featured in this tree to their containing group and the rest of the Tree of Life. The basal branching point in the tree represents the ancestor of the other groups in the tree. This ancestor diversified over time into several descendent subgroups, which are represented as internal nodes and terminal taxa to the right.

You can click on the root to travel down the Tree of Life all the way to the root of all Life, and you can click on the names of descendent subgroups to travel up the Tree of Life all the way to individual species.
For more information on ToL tree formatting, please see Interpreting the Tree or Classification. To learn more about phylogenetic trees, please visit our Phylogenetic Biology pages.
close boxPhylogenetic tree based on analyses of ribosomal small subunit sequences modified from Suh et al., 2006.
Introduction
The ascomycete yeasts (phylum Ascomycota: subphylum Saccharomycotina: class Saccharomycetes: order Saccharomycetales) constitute a monophyletic group of economically and environmentally important fungi. Most people know that ascomycete yeasts produce CO2 that makes bread rise and ferment sugary substrates to alcohol. In fact many yeast species are vitally important in modern commerce.
Some species are parasites of animals and plants. Candida albicans and its close relatives cause disease in human beings, and for this reason the C. albicans genome was one of the first yeast genomes sequenced. Although we live surrounded by this yeast, only individuals with compromised immune systems are at risk for developing invasive candidiasis. Invasive candidiasis is the fourth most common bloodstream infection in hospitalized patients, and the U. S. Centers for Disease Control and Prevention report that eight out of every 100,000 persons develop blood infections every year. Such infections in immunocompromized individuals may result in death if antibiotic therapy is not effective.
Eremothecium gossypii and its relatives are filamentous plant pathogenic yeasts transmitted by sucking insects including stinkbugs. In addition to cotton (Gossypium sp., hence the species name) some fruits, including citrus, also are infected resulting in soft brown rot. This yeast also produces copious amounts of riboflavin (vitamin B2) that apparently protects the yeast spores against ultraviolet light as well as serving as a commercial source of the vitamin. The complete genome of E. gossypii has been sequenced, indicating its importance as a model organism.
Many more ascomycete yeasts, however, are free-living in substrates of high organic content in close associations with other organisms. There are many examples of their interactions with arthropods, and yeasts may provide vitamins and enzymes to arthropods in exchange for efficient habitat and dispersal. About 1000 species of Saccharomycetales are known, and based on species accumulation curves in ongoing surveys of yeasts at many localities, it is estimated that large numbers of taxa remain to be discovered that will double the known number.
Characteristics
‘Yeast’ describes a growth form that is not restricted to this group of ascomycetes, but rather refers to the fact that the fungal body is single celled and reproduces asexually by budding. In Saccharomycetales somatic cells convert to free asci during sexual reproduction and are not enclosed in ascomata (fruiting bodies). Many yeasts, especially basal members of the group, may have an extensive mycelium. Several ultrastructural characters distinguish these yeasts from most other ascomycetes. In meiosis the spindles of the two successive divisions occur within the intact nuclear envelop with the spindles oriented perpendicular to each other. In electron microscope observations ascospore delimiting membranes do not form a common sac around the nuclei after meiosis, but rather, first appear separately associated with individual nuclei. In addition Saccharomycetales appear to lack Woronin bodies, organelles that have a role in plugging septal pores, although some of the Taphrinomycotina also may lack Woronin bodies.
Ascomycete yeasts differ from other yeast growth forms, particularly basidiomycete yeasts, by other traits including lower amounts of chitin in their cell walls, nuclear DNA with relatively low guanine + cytosine content, holoblastic bud formation with wall layers remaining continuous, often with fermentative ability, and, usually, inability to stain with diazonium blue. Some yeastlike ascomycetes that are recently derived from filamentous yeasts (Pezizomycotina) are easily distinguished by DNA comparison.
Reproduction and Life Cycle
The life cycle of Saccharomyces cerevisiae has been studied extensively, and because its entire genome was sequenced (the first to be completed in eukaryotes), reproductive processes also are well known at the molecular level. In asexual reproduction the cell nucleus divides by mitosis and as a bud develops a second nucleus moves into the cell. The bud enlarges and at the point when it reaches the size of the parent cell it separates from it. The majority of the nearly 1000 known ascomycete yeasts are believed to reproduce only asexually, but it often is difficult to detect sexual reproduction in newly discovered heterothallic species unless adequate numbers of different isolates are crossed in an attempt to discover both mating types.

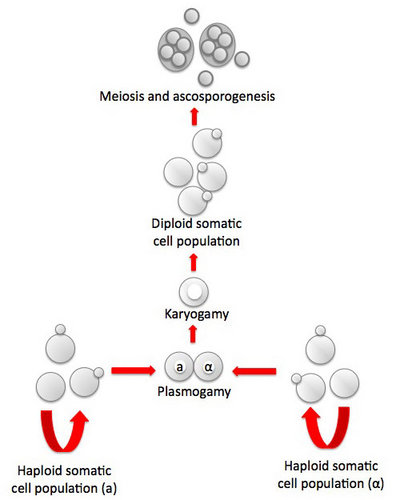
Fig. 1. Life cycle of Saccharomyces cerevisiae. Asexual reproduction occurs by budding in both haploid and diploid populations of cells. In sexual reproduction attraction by pheromone signaling by haploid cells is followed by plasmogamy and karyogamy. Meiosis may be delayed until after a population of diploid cells is produced. Meiosis and ascosporogenesis result in the production of haploid ascospores. Ascospores are not produced in an ascoma (fruiting body), one hallmark of the ascomycete yeasts. © Meredith Blackwell
In Saccharomyces cerevisiae sexual reproduction occurs after cells containing different mating type alleles designated a and α fuse in response to mating pheromones in a process called plasmogamy. Karyogamy, the fusion of nuclei and the second step in the process of fertilization, occurs to yield a cell with a diploid nucleus. There may, however, be a delay in the completion of sexual reproduction, and the population of budding diploid cells may increase in number. Eventually the mated cells develop into asci and undergo meiosis. As mentioned in the discussion of the characters of ascomycete yeasts, the nuclear membrane remains intact during both meiosis I and II, and the spindle of the second division is parallel to that of the first division. Cleavage of four ascospores is initiated by ascospore delimiting membranes that develop independently in association with each of the four nuclei that are the products of meiosis. The ascospores are passively released from the ascus. In contrast to all members of the Pezizomycotina and to Neolecta (Taphrinomycotina), the asci of S. cerevisiae and other yeasts are never enclosed in an ascoma (fruiting body).
Ecology and Physiology
Members of Saccharomycetales generally occupy damp or wet habitats that are high in organic material. Clades often but not always can be defined by their ecological traits. S. cerevisiae clade members are not well known in nature. They occur with regularity in the bark of certain deciduous trees (Sampaio and Gonçalves, 2008) and intermittently in fermenting fruit and other high sugar environments such as nectar and sap fluxes. Members of the clade have high tolerance to alcohol. Many of these yeasts grow anaerobically and ferment glucose to alcohol.
Other clades are associated with necrotic tissues of succulent plants, brine, mushrooms and other fungi, and partially rotted wood. Yeasts of the Candida albicans clade form invasive hyphae that grow at 37ºC (human body temperature) contributing to their ability to infect humans and other mammals. Some yeasts are closely associated with herbivorous arthropods. These include species of Metschnikowia associated with beetles and other insects that visit flowers (Lachance et al., 2001). Others, primarily in the Candida tanzawaensis clade, are associated with mushroom-feeding beetles.
There is growing evidence that the vast majority of ascomycete yeasts are involved with insects and other arthropods. A classic example of yeast-insect interactions involves cactophilic yeasts and species of Drosophila that are found in the decaying cacti native to the Sonoran Desert. The flies are vectors of the yeasts that they inoculate into necrotic lesions of the cacti, and heterogenous yeast assemblages may be important as fly attractants (Starmer et al., 1990).
In addition to the yeast-beetle associations mentioned above, several members of the Scheffersomyces stipitis clade are involved with wood-ingesting insects in a variety of families and even orders. The clade members have in common that they ferment xylose, a rare trait among yeasts. It is not clear if the insect benefits from the association, but in a striking example every one of 400 individual passalid beetles had S. stipitis in its gut (unpublished, Blackwell and Nguyen). The yeast may simply occupy a xylose-rich niche to its own advantage, because it is located at the extreme posterior end of the beetle's hindgut where there normally is little absorption from the gut. Species of Starmerella are found with bees and Wickerhamiella with drosophilas that visit flowers (Lachance et al., 2001).
Another interesting clade centered on the genus Saccharomycopsis, consists of about fifteen species, most of which are predaceous (Fig. 2, Lachance et al., 2000). The cells develop infection pegs that are used to invade and kill other yeasts under certain conditions. A variety of both basidiomycete and ascomycete yeasts can be penetrated by the predators. Environmental conditions vary for predation and rich nitrogenous nutrients, organic sulfur compounds, or high concentrations of ammonium nitrogen may inhibit or promote predation, depending on the yeast. Predaceous species may attack each other.

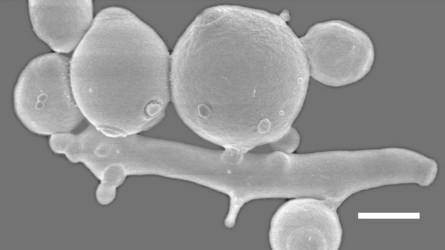
Fig. 2. Scanning electron micrograph showing of elongated cells of Arthroascus schoenii with infection pegs penetrating spherical cells of Saccharomyces cerevisiae. Bar = 2µm. Image by B. Schlag-Edler, A. Pupovac-Velikonja and M.-A. Lachance, from Suh et al. 2006, © The Mycological Society of America.
Recent findings indicate another relationship between yeasts and vertebrates. Tree shrews and other small tropical mammals consume alcohol produced by yeasts in the special flower buds of a palm tree, an association that has endured for at least 55 million years (Wiens et al., 2008).
Biogeography
A large amount of research has shown that most yeasts associated with natural habitats are restricted in their distributions. In an extensive study of yeasts dispersed to ephemeral flowers such as morning glories by insect vectors, some yeast species have been found to be endemic to specific regions, such as North, Central, or South America. Others are native to Asia and Australia or exclusive to the Hawaiian Islands (Lachance et al., 2001, 2005). Many other examples can be found among yeasts associated with fungus-feeding beetles in families Erotylidae and Nitulidiae, some of which appear to have specificity for beetles with restricted distributions.
Although some yeasts may have naturally broad distributions because they are air dispersed, there is evidence that others have been dispersed by human activities. At least one species, Clavispora opuntiae, has low DNA sequence variation in localities distant to its probable origin in Argentina. Apparently, the yeast traveled with an Argentinean moth, Cactoblastis cactorum, spread globally in an attempt to control cacti introduced into Australia, Hawaii, and the Caribbean. Two Metschnikowia species from Central America were apparently introduced to Hawaii through plant material infested with beetles. The species have high degrees of genetic diversity in their native land, but almost none in their new territory (Lachance et al., 2008).
Discussion of Phylogenetic Relationships
Members of the subphylum Saccharomycotina constitute a monophyletic group of ascomycetes that are well defined by ultrastructural and DNA characteristics. Saccharomycotina have been well established in numerous studies of ascomycetes as the sister group to Pezizomycotina. The basal ascomycete group, Taphrinomycotina, is the sister to Saccharomycotina and Pezizomycotina. Although members of Saccharomycotina were considered to be primitive by some early mycologists, they are best viewed as a highly divergent group from the sister taxon. The subphylum contains a single class and order (Saccharomycetes and Saccharomycetales).
Early yeast classification involved the defining of a monophyletic group in the absence of DNA analysis. The classification of yeasts as a monophyletic group meant removing the basidiomycetes following the discovery of clamps and basidiospores in some species (Kurtzman and Fell 1998). As discussed above (see Characteristics) several physiological characters also were useful. DNA sequence analysis allowed the transfer of yeast forms now placed in Taphrinomycotina (e.g., Neolecta, Schizosaccharomyces) and Pezizomycotina (e.g., Symbiotaphrina, yeast-like symbionts of plant hoppers) to their correct phylogenetic position.
An understanding of the relationships among species of Saccharomycetales requires additional work. In particular, more taxa need to be sampled using a multilocus database. Although there are several dozen full yeast genomes available, the sampling has not been even throughout the taxa and basal clades have not been sampled fully.
A number of clades, however, are well supported by available molecular data, and these often are supported further by morphological and physiological traits (see above, Ecology and Physiology). Taxonomy generally lags behind phylogenetic studies, and all family level taxa are not yet well established. Several clades of interest, however, are depicted in the phylogenetic tree. The more basal clade members (e.g., Blastobotrys terrestris, Lipomyces oligophaga) often are mycelial but with some budding cells. Yeasts in the Saccharomyces cerevisiae clade used in baking and brewing are among the derived yeast taxa. Several yeasts in the Candida albicans clade cause disease in mammals. Other notable yeasts include members of the Scheffersomyces stipitis clade with the unusual ability to ferment xylose to alcohol; and the Candida tanzawaensis and Pichia membranifaciens clades, among those yeasts often associated with insects.
Classification
Because yeasts have little morphological variation, they were traditionally classified not only by morphology but also physiology. It should be noted that other fungi are called yeasts. The basidiomycete yeasts have budding cells and a basidial state that is not enclosed in a fruiting body, parallel with ascomycete yeasts that also have budding cells and asci not enclosed in a fruiting body.
The ability to determine their phylogenetic relationships with some certainty, however, has come only recently with the advent of DNA sequencing (Kurtzman and Robnett, 2003, 2007; Kurtzman et al. 2007, 2008). In fact yeasts were one of the first groups of organisms to have all taxa sequenced in a specific region, so that rapid identification became possible very early (Kurtzman and Robnett, 1998). This process is sometimes known as barcoding. Although many yeasts were sequenced, multigene sequence datasets only recently became available for Saccharomycetales, and within the order families are not yet fully resolved. It is evident that despite a large number of full genomes known for yeasts, many of the basal yeasts are less well known.
One point to clarify is the use of the term “yeast” by many biologists to indicate the species known as S. cerevisiae. We reemphasize that there are about 1000 known species, and one of them is S. cerevisiae, and it is among the species that several problems in classification arise. Many taxa that are not known to produce a sexual state have been lumped into Candida, and the process of assigning phylogenetically relevant names is still underway. In another example, taxa that produce a hat-shaped ascospore were once included as species of Pichia. Once the generic name changes are complete, the classification will be greatly improved. Also, although a number of clades are well supported as we already mentioned, a complete phylogenetic classification cannot yet be achieved because of incomplete sampling. In particular, taxonomic groupings above genus have not always been reliably established.
References
Hibbett, D. S., M. Binder, J. F. Bischoff, M. Blackwell, P. F. Cannon, O. E. Eriksson, S. Huhndorf, T. James, P. M. Kirk, R. Lücking, T. Lumbsch, F. Lutzoni, P. B. Matheny, D. J. Mclaughlin, M. J. Powell, S. Redhead, C. L. Schoch, J. W. Spatafora, J. A. Stalpers, R. Vilgalys, M. C. Aime, A. Aptroot, R. Bauer, D. Begerow, G. L. Benny, L. A. Castlebury, P. W. Crous, Y.-C. Dai, W. Gams, D. M. Geiser, G. W. Griffith, C. Gueidan, D. L. Hawksworth, G. Hestmark, K. Hosaka, R. A. Humber, K. Hyde, J. E. Ironside, U. Kõljalg, C. P. Kurtzman, K.-H. Larsson, R. Lichtwardt, J. Longcore, J. Miądlikowska, A. Miller, J.-M. Moncalvo, S. Mozley-Standridge, F. Oberwinkler, E. Parmasto, V. Reeb, J. D. Rogers, C. Roux, L. Ryvarden, J. P. Sampaio, A. Schüßler, J. Sugiyama, R. G. Thorn, L. Tibell, W. A. Untereiner, C. Walker, Z. Wang, A. Weir, M. Weiß, M. M. White, K. Winka, Y.-J. Yao, and N. Zhang. 2007. A higher-level phylogenetic classification of the Fungi. Mycological Research 111: 509-547.
Kurtzman, C.P., J. Albertyn and E. Basehoar-Powers. 2007. Multigene phylogenetic analysis of the Lipomycetaceae and the proposed transfer of Zygozyma species to Lipomyces and Babjevia anomala to Dipodascopsis. FEMS Yeast Res. 7, 1027-1034.
Kurtzman, C. P., J. W. Fell, eds. 1998. The yeasts, a taxonomic study, 4th ed. Amsterdam: Elsevier. 1055 p.
Kurtzman, C. P. and C. J. Robnett. 1998. Identification and phylogeny of
ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73:331–371.
Kurtzman, C.P., and C.J. Robnett. 2003. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res 3, 417-432.
Kurtzman, C.P. and C.J. Robnett. 2007. Multigene phylogenetic analysis of the Trichomonascus, Wickerhamiella and Zygoascus yeast clades, and the proposal of Sugiyamaella gen. nov and 14 new species combinations. FEMS Yeast Res. 7, 141-151.
Kurtzman, C.P., C.J. Robnett and E. Basehoar-Powers. 2008. Relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene phylogenetic analysis and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS Yeast Res. In press.
Kurtzman, C. P. and M. Suzuki. 2008. Phylogenetic analysis of ascomycete yeasts that form coenzyme Q-9 and the proposal of the new genera Babjeviella, Meyerozyma, Millerozyma, Priceomyces and Scheffersomyces. Mycoscience. In press.
Lachance M. A., C. P. Ewing, J. M. Bowles and W. T Starmer. 2005. Metschnikowia hamakuensis sp. nov., Metschnikowia kamakouana sp. nov., and Metschnikowia mauinuiana sp. nov., three endemic yeasts from Hawaiian nitidulid beetles. Int. J. Syst. Evol. Microbiol. 55:1369-1377.
Lachance M. A., D. Lawrie, J. Dobson, and J. Piggott. 2008. Biogeography and population structure of the Neotropical endemic yeast species Metschnikowia lochheadii. Antonie van Leeuwenhoek 94:403-414.
Lachance, M. A, A. Pupovac-Velikonja, S. Natarajan, and B. Schlag-Edler. 2000. Nutrition and phylogeny of predacious yeasts. Can. J. Microbiol. 46: 495–505.
Lachance, M. A., W. T. Starmer, J. M. Bowles, H. J. Phaff, and C. A. Rosa. 2000. Ribosomal DNA, species structure, and biogeography of the cactophilic yeast Clavispora opuntiae Can. J. Microbiol. Mar, 46:195–210.
Lachance M. A., W. T. Starmer, C. A. Rosa, J. M. Bowles, J. S. F. Barker, and D. H. Janzen. 2001. Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Res. 1:1-8
Sampaio, J. P. and P. Gonçalves. 2008. Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and are sympatric with S. cerevisiae and S. paradoxus. Appl. Environ. Microbiol. 74:2144–2152
Starmer, W. T., M. A. Lachance, H. J. Phaff, and W. B. Heed. 1990. The biogeography of yeasts associated with cactus tissue in North America, the Caribbean and northern Venezuela. Evol. Biol. 24:253-296.
Suh, S.-O., M. Blackwell, C. P. Kurtzman, and M-A. Lachance. 2006. Phylogenetics of Saccharomycetales, the ascomycete yeasts. Mycologia 98:1008-1019.
Wiens F., A. Zitzmann, M. A. Lachance, M. Yegles, F. Pragst, F. M. Wurst, D. von Holst, S. L. Guan, and R. Spanagel. 2008. Chronic intake of fermented floral nectar by wild treeshrews. Proc Nat Acad Sci 105:10426-10431.
Title Illustrations

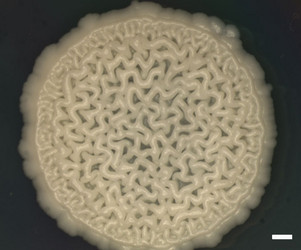
| Scientific Name | Y-48049 (undescribed) |
|---|---|
| Comments | Colony of undescribed yeast taxon (Y-48049) from the gut of a fungus-eating beetle. A wrinkled colony surface is characteristic of some yeast species. Note that the yeast colony (Y-48049) shows restricted growth on agar media. |
| Acknowledgements | Christine Ackerman |
| Size | Bar = 1 mm |
| Collection | Meredith Blackwell |
| Copyright |
© Meredith Blackwell

|
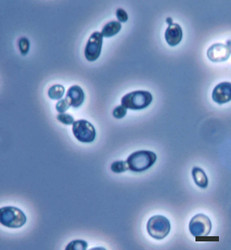
| Scientific Name | Saccharomyces cerevisiae |
|---|---|
| Comments | Brewer's yeast, Saccharomyces cerevisiae (Y-2235) cells; note many budding cells. |
| Size | Bar = 5µm |
| Copyright |
©

|
| Scientific Name | Kodamaea anthophila |
|---|---|
| Comments | Scanning electron microscope image of two hat-shaped ascospores of Kodamaea anthophila; This yeast species is dispersed between ephemeral flowers, including Hibiscus spp., by beetles. |
| Reference | Rosa, C.A. M.A. Lachance, W.T. Starmer, J.F. Barker, J.M. Bowles, and B. Schlag Edler. 1999. Kodamaea nitidulidarum, Candida restingae, and Kodamaea anthophila, three new related yeast species from ephemeral flowers. Int. J. Syst. Bacteriol. 49:309 318. |
| Size | Bar = 2 µm |
| Copyright | © 1999 Society for General Microbiology |
About This Page
Meredith Blackwell

Louisiana State University, Baton Rouge, Louisiana, USA

National Center for Agricultural Utilization Research, Peoria, Illinois, USA
Marc-André Lachance

University of Western Ontario, London, Ontario, Canada
Sung-Oui Suh

American Type Culture Collection (ATCC), Manassas, Virginia, USA
Correspondence regarding this page should be directed to Meredith Blackwell at , Cletus P. Kurtzman at , Marc-André Lachance at , and Sung-Oui Suh at
Page copyright © 2009 Meredith Blackwell, , Marc-André Lachance, and Sung-Oui Suh
All Rights Reserved.
- First online 09 January 2008
- Content changed 22 January 2009
Citing this page:
Blackwell, Meredith, Cletus P. Kurtzman, Marc-André Lachance, and Sung-Oui Suh. 2009. Saccharomycotina. Saccharomycetales. Version 22 January 2009. http://tolweb.org/Saccharomycetales/29043/2009.01.22 in The Tree of Life Web Project, http://tolweb.org/








 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site